Channel Quality Assessment, Pruning, and Motion Artifact Detection
This notebook sketches how to prune bad channels and detect motion artefacts in fNIRS data
[1]:
import cedalion
import cedalion.nirs
import cedalion.sigproc.quality as quality
import cedalion.xrutils as xrutils
import cedalion.datasets as datasets
import xarray as xr
import matplotlib.pyplot as p
from cedalion import units
Loading raw CW-NIRS data from a SNIRF file and converting it to OD and CONC
This notebook uses a finger-tapping dataset in BIDS layout provided by Rob Luke that is automatically fetched. You can also find it here.
[2]:
# get example finger tapping dataset
rec = datasets.get_fingertapping()
rec["od"] = cedalion.nirs.int2od(rec["amp"])
# Plot some data for visual validation
f,ax = p.subplots(1,1, figsize=(12,4))
ax.plot( rec["amp"].time, rec["amp"].sel(channel="S3D3", wavelength="850"), "r-", label="850nm")
ax.plot( rec["amp"].time, rec["amp"].sel(channel="S3D3", wavelength="760"), "r-", label="760nm")
p.legend()
ax.set_xlabel("time / s")
ax.set_ylabel("Signal intensity / a.u.")
display(rec["amp"])
<xarray.DataArray (channel: 28, wavelength: 2, time: 23239)> Size: 10MB
<Quantity([[[0.0913686 0.0909875 0.0910225 ... 0.0941083 0.0940129 0.0944882]
[0.1856806 0.186377 0.1836514 ... 0.1856486 0.1850836 0.1842172]]
[[0.227516 0.2297024 0.2261366 ... 0.2264519 0.2271665 0.226713 ]
[0.6354927 0.637668 0.6298023 ... 0.6072068 0.6087293 0.6091066]]
[[0.1064704 0.1066212 0.1053444 ... 0.121114 0.1205022 0.1205441]
[0.2755033 0.2761615 0.2727006 ... 0.2911952 0.2900544 0.2909847]]
...
[[0.2027881 0.1996586 0.2004866 ... 0.2318743 0.2311941 0.2330808]
[0.4666358 0.4554404 0.4561614 ... 0.4809749 0.4812827 0.4862896]]
[[0.4885007 0.4802285 0.4818338 ... 0.6109142 0.6108118 0.613845 ]
[0.8457658 0.825988 0.8259648 ... 0.975894 0.9756599 0.9826459]]
[[0.6304559 0.6284427 0.6287045 ... 0.6810626 0.6809573 0.6818709]
[1.2285622 1.2205907 1.2190002 ... 1.2729124 1.2727222 1.2755645]]], 'volt')>
Coordinates:
* time (time) float64 186kB 0.0 0.128 0.256 ... 2.974e+03 2.974e+03
samples (time) int64 186kB 0 1 2 3 4 5 ... 23234 23235 23236 23237 23238
* channel (channel) object 224B 'S1D1' 'S1D2' 'S1D3' ... 'S8D8' 'S8D16'
source (channel) object 224B 'S1' 'S1' 'S1' 'S1' ... 'S8' 'S8' 'S8'
detector (channel) object 224B 'D1' 'D2' 'D3' 'D9' ... 'D7' 'D8' 'D16'
* wavelength (wavelength) float64 16B 760.0 850.0
Attributes:
data_type_group: unprocessed raw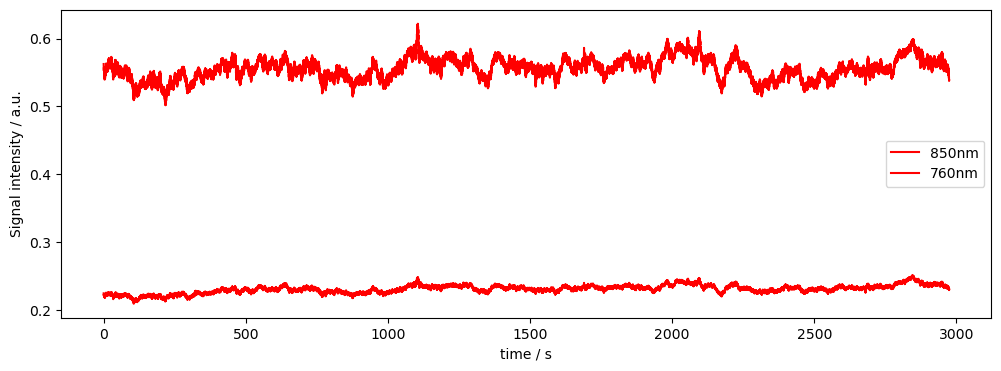
Calculating Signal Quality Metrics and applying Masks
To assess channel quality metrics such as SNR, channel distances, average amplitudes, sci, and others, we use small helper functions. As input, the quality functions should also expect thresholds for these metrics, so they can feed back both the calculated quality metrics as well as a mask. The masks can then be combined and applied - e.g. to prune channels with low SNR. The input and output arguments are based on xarray time series, quality parameters / instructions for thresholding. The returned mask is a boolean array in the shape and size of the input time series. It indicates where the threshold for our quality metric was passed (“True”) and is False otherwise. Masks can be combined with other masks, for instance to apply several metrics to assess or prune channels. At any point in time, the mask can be applied using the “apply_mask()” function available from cedalion’s the xrutils package.
If you are a user who is mainly interested in high-level application, you can skip to the Section “Channel Pruning using Quality Metrics and the Pruning Function” below. The “prune_ch()” function provides a higher abstraction layer to simply prune your data, using the same metrics and functions that are demonstrated below.
Channel Quality Metrics: SNR
[3]:
# Here we assess channel quality by SNR
snr_thresh = 16 # the SNR (std/mean) of a channel. Set high here for demonstration purposes
# SNR thresholding using the "snr" function of the quality subpackage
snr, snr_mask = quality.snr(rec["amp"], snr_thresh)
# apply mask function. In this example, we want all signals with an SNR below the threshold to be replaced with "nan".
# We do not want to collapse / combine any dimension of the mask (last argument: "none")
data_masked_snr_1, masked_elements_1 = xrutils.apply_mask(rec["amp"], snr_mask, "nan", "none")
# alternatively, we can "drop" all channels with an SNR below the threshold. Since the SNR of both wavelength might differ
# (pass the threshold for one wavelength, but not for the other), we collapse to the "channel" dimension.
data_masked_snr_2, masked_elements_2 = xrutils.apply_mask(rec["amp"], snr_mask, "drop", "channel")
# show some results
print(f"channels that were masked according to the SNR threshold: {masked_elements_2}")
# dropped:
data_masked_snr_2
mask collapsed to channel dimension
channels that were masked according to the SNR threshold: ['S4D4' 'S5D7' 'S6D8' 'S8D8']
[3]:
<xarray.DataArray (channel: 24, wavelength: 2, time: 23239)> Size: 9MB
<Quantity([[[0.0913686 0.0909875 0.0910225 ... 0.0941083 0.0940129 0.0944882]
[0.1856806 0.186377 0.1836514 ... 0.1856486 0.1850836 0.1842172]]
[[0.227516 0.2297024 0.2261366 ... 0.2264519 0.2271665 0.226713 ]
[0.6354927 0.637668 0.6298023 ... 0.6072068 0.6087293 0.6091066]]
[[0.1064704 0.1066212 0.1053444 ... 0.121114 0.1205022 0.1205441]
[0.2755033 0.2761615 0.2727006 ... 0.2911952 0.2900544 0.2909847]]
...
[[0.187484 0.1868235 0.1866562 ... 0.1735965 0.1736705 0.1738339]
[0.2424386 0.241503 0.2408491 ... 0.22303 0.2229887 0.2234081]]
[[0.2027881 0.1996586 0.2004866 ... 0.2318743 0.2311941 0.2330808]
[0.4666358 0.4554404 0.4561614 ... 0.4809749 0.4812827 0.4862896]]
[[0.6304559 0.6284427 0.6287045 ... 0.6810626 0.6809573 0.6818709]
[1.2285622 1.2205907 1.2190002 ... 1.2729124 1.2727222 1.2755645]]], 'volt')>
Coordinates:
* time (time) float64 186kB 0.0 0.128 0.256 ... 2.974e+03 2.974e+03
samples (time) int64 186kB 0 1 2 3 4 5 ... 23234 23235 23236 23237 23238
* channel (channel) object 192B 'S1D1' 'S1D2' 'S1D3' ... 'S8D7' 'S8D16'
source (channel) object 192B 'S1' 'S1' 'S1' 'S1' ... 'S7' 'S8' 'S8'
detector (channel) object 192B 'D1' 'D2' 'D3' 'D9' ... 'D15' 'D7' 'D16'
* wavelength (wavelength) float64 16B 760.0 850.0
Attributes:
data_type_group: unprocessed rawChannel Quality Metrics: Channel Distance
[4]:
# Here we assess channel distances. We might want to exclude very short or very long channels
sd_threshs = [1, 4.5]*units.cm # defines the lower and upper bounds for the source-detector separation that we would like to keep
# Source Detector Separation thresholding
ch_dist, sd_mask = quality.sd_dist(rec["amp"], rec.geo3d, sd_threshs)
# print the channel distances
print(f"channel distances are: {ch_dist}")
# apply mask function. In this example, we want to "drop" all channels that do not fall inside sd_threshs
# i.e. drop channels shorter than 1cm and longer than 4.5cm. We want to collapse along the "channel" dimension.
data_masked_sd, masked_elements = xrutils.apply_mask(rec["amp"], sd_mask, "drop", "channel")
# display the resultings
print(f"channels that were masked according to the SD Distance thresholds: {masked_elements}")
data_masked_sd
channel distances are: <xarray.DataArray (channel: 28)> Size: 224B
<Quantity([0.039 0.039 0.041 0.008 0.037 0.038 0.037 0.007 0.04 0.037 0.008 0.041
0.034 0.008 0.039 0.039 0.041 0.008 0.037 0.037 0.037 0.008 0.04 0.037
0.007 0.041 0.033 0.008], 'meter')>
Coordinates:
* channel (channel) object 224B 'S1D1' 'S1D2' 'S1D3' ... 'S8D8' 'S8D16'
source (channel) object 224B 'S1' 'S1' 'S1' 'S1' ... 'S7' 'S8' 'S8' 'S8'
detector (channel) object 224B 'D1' 'D2' 'D3' 'D9' ... 'D7' 'D8' 'D16'
mask collapsed to channel dimension
channels that were masked according to the SD Distance thresholds: ['S1D9' 'S2D10' 'S3D11' 'S4D12' 'S5D13' 'S6D14' 'S7D15' 'S8D16']
[4]:
<xarray.DataArray (channel: 20, wavelength: 2, time: 23239)> Size: 7MB
<Quantity([[[0.0913686 0.0909875 0.0910225 ... 0.0941083 0.0940129 0.0944882]
[0.1856806 0.186377 0.1836514 ... 0.1856486 0.1850836 0.1842172]]
[[0.227516 0.2297024 0.2261366 ... 0.2264519 0.2271665 0.226713 ]
[0.6354927 0.637668 0.6298023 ... 0.6072068 0.6087293 0.6091066]]
[[0.1064704 0.1066212 0.1053444 ... 0.121114 0.1205022 0.1205441]
[0.2755033 0.2761615 0.2727006 ... 0.2911952 0.2900544 0.2909847]]
...
[[0.2225884 0.2187791 0.2195495 ... 0.2564863 0.2551258 0.2560233]
[0.3994258 0.3917637 0.389261 ... 0.4304597 0.430814 0.4331249]]
[[0.2027881 0.1996586 0.2004866 ... 0.2318743 0.2311941 0.2330808]
[0.4666358 0.4554404 0.4561614 ... 0.4809749 0.4812827 0.4862896]]
[[0.4885007 0.4802285 0.4818338 ... 0.6109142 0.6108118 0.613845 ]
[0.8457658 0.825988 0.8259648 ... 0.975894 0.9756599 0.9826459]]], 'volt')>
Coordinates:
* time (time) float64 186kB 0.0 0.128 0.256 ... 2.974e+03 2.974e+03
samples (time) int64 186kB 0 1 2 3 4 5 ... 23234 23235 23236 23237 23238
* channel (channel) object 160B 'S1D1' 'S1D2' 'S1D3' ... 'S8D7' 'S8D8'
source (channel) object 160B 'S1' 'S1' 'S1' 'S2' ... 'S7' 'S8' 'S8'
detector (channel) object 160B 'D1' 'D2' 'D3' 'D1' ... 'D7' 'D7' 'D8'
* wavelength (wavelength) float64 16B 760.0 850.0
Attributes:
data_type_group: unprocessed rawChannel Quality Metrics: Mean Amplitudes
[5]:
# Here we assess average channel amplitudes. We might want to exclude very small or large signals
amp_threshs = [0.1, 3]*units.volt # define whether a channel's amplitude is within a certain range
# Amplitude thresholding
mean_amp, amp_mask = quality.mean_amp(rec["amp"], amp_threshs)
# apply mask function. In this example, we want drop all channels that do not fall inside the amplitude thresholds.
# We collapse to the "channel" dimension.
data_masked_amp, masked_elements = xrutils.apply_mask(rec["amp"], amp_mask, "drop", "channel")
# display the results
print(f"channels that were masked according to the amplitude threshold: {masked_elements}")
data_masked_amp
mask collapsed to channel dimension
channels that were masked according to the amplitude threshold: ['S1D1' 'S1D9' 'S3D2' 'S6D8' 'S7D6']
[5]:
<xarray.DataArray (channel: 23, wavelength: 2, time: 23239)> Size: 9MB
<Quantity([[[0.227516 0.2297024 0.2261366 ... 0.2264519 0.2271665 0.226713 ]
[0.6354927 0.637668 0.6298023 ... 0.6072068 0.6087293 0.6091066]]
[[0.1064704 0.1066212 0.1053444 ... 0.121114 0.1205022 0.1205441]
[0.2755033 0.2761615 0.2727006 ... 0.2911952 0.2900544 0.2909847]]
[[0.5512474 0.5510672 0.5476283 ... 0.6179242 0.6188702 0.6187721]
[1.125532 1.1238331 1.1119423 ... 1.1817728 1.1819598 1.1832658]]
...
[[0.2027881 0.1996586 0.2004866 ... 0.2318743 0.2311941 0.2330808]
[0.4666358 0.4554404 0.4561614 ... 0.4809749 0.4812827 0.4862896]]
[[0.4885007 0.4802285 0.4818338 ... 0.6109142 0.6108118 0.613845 ]
[0.8457658 0.825988 0.8259648 ... 0.975894 0.9756599 0.9826459]]
[[0.6304559 0.6284427 0.6287045 ... 0.6810626 0.6809573 0.6818709]
[1.2285622 1.2205907 1.2190002 ... 1.2729124 1.2727222 1.2755645]]], 'volt')>
Coordinates:
* time (time) float64 186kB 0.0 0.128 0.256 ... 2.974e+03 2.974e+03
samples (time) int64 186kB 0 1 2 3 4 5 ... 23234 23235 23236 23237 23238
* channel (channel) object 184B 'S1D2' 'S1D3' 'S2D1' ... 'S8D8' 'S8D16'
source (channel) object 184B 'S1' 'S1' 'S2' 'S2' ... 'S8' 'S8' 'S8'
detector (channel) object 184B 'D2' 'D3' 'D1' 'D3' ... 'D7' 'D8' 'D16'
* wavelength (wavelength) float64 16B 760.0 850.0
Attributes:
data_type_group: unprocessed rawChannel Pruning using Quality Metrics and the Pruning Function
To prune channels according to quality criteria, we do not have to manually go through the steps above. Instead, we can create quality masks for the metrics that we are interested in and hand them to a dedicated channel pruning function. The prune function expects a list of quality masks alongside a logical operator that defines how these masks should be combined.
[6]:
# as above we use three metrics and define thresholds accordingly
snr_thresh = 16 # the SNR (std/mean) of a channel.
sd_threshs = [1, 4.5]*units.cm # defines the lower and upper bounds for the source-detector separation that we would like to keep
amp_threshs = [0.1, 3]*units.volt # define whether a channel's amplitude is within a certain range
# then we calculate the masks for each metric: SNR, SD distance and mean amplitude
_, snr_mask = quality.snr(rec["amp"], snr_thresh)
_, sd_mask = quality.sd_dist(rec["amp"], rec.geo3d, sd_threshs)
_, amp_mask = quality.mean_amp(rec["amp"], amp_threshs)
# put all masks in a list
masks = [snr_mask, sd_mask, amp_mask]
# prune channels using the masks and the operator "all", which will keep only channels that pass all three metrics
amp_pruned, drop_list = quality.prune_ch(rec["amp"], masks, "all")
# print list of dropped channels
print(f"List of pruned channels: {drop_list}")
# display the new data xarray
amp_pruned
mask collapsed to channel dimension
List of pruned channels: ['S1D1' 'S1D9' 'S2D10' 'S3D2' 'S3D11' 'S4D4' 'S4D12' 'S5D7' 'S5D13' 'S6D8'
'S6D14' 'S7D6' 'S7D15' 'S8D8' 'S8D16']
[6]:
<xarray.DataArray (channel: 13, wavelength: 2, time: 23239)> Size: 5MB
<Quantity([[[0.227516 0.2297024 0.2261366 ... 0.2264519 0.2271665 0.226713 ]
[0.6354927 0.637668 0.6298023 ... 0.6072068 0.6087293 0.6091066]]
[[0.1064704 0.1066212 0.1053444 ... 0.121114 0.1205022 0.1205441]
[0.2755033 0.2761615 0.2727006 ... 0.2911952 0.2900544 0.2909847]]
[[0.5512474 0.5510672 0.5476283 ... 0.6179242 0.6188702 0.6187721]
[1.125532 1.1238331 1.1119423 ... 1.1817728 1.1819598 1.1832658]]
...
[[0.3463254 0.3424951 0.3408207 ... 0.3929267 0.3941368 0.3945422]
[0.6978315 0.6875081 0.6857653 ... 0.7259991 0.7271688 0.7292138]]
[[0.2225884 0.2187791 0.2195495 ... 0.2564863 0.2551258 0.2560233]
[0.3994258 0.3917637 0.389261 ... 0.4304597 0.430814 0.4331249]]
[[0.2027881 0.1996586 0.2004866 ... 0.2318743 0.2311941 0.2330808]
[0.4666358 0.4554404 0.4561614 ... 0.4809749 0.4812827 0.4862896]]], 'volt')>
Coordinates:
* time (time) float64 186kB 0.0 0.128 0.256 ... 2.974e+03 2.974e+03
samples (time) int64 186kB 0 1 2 3 4 5 ... 23234 23235 23236 23237 23238
* channel (channel) object 104B 'S1D2' 'S1D3' 'S2D1' ... 'S7D7' 'S8D7'
source (channel) object 104B 'S1' 'S1' 'S2' 'S2' ... 'S6' 'S7' 'S8'
detector (channel) object 104B 'D2' 'D3' 'D1' 'D3' ... 'D7' 'D7' 'D7'
* wavelength (wavelength) float64 16B 760.0 850.0
Attributes:
data_type_group: unprocessed rawMotion Artefact Detection
The same xarray-based masks can be used for indicating motion-artefacts. The example below shows how to checks channels for motion artefacts using standard thresholds from Homer2/3. The output is a mask that can be handed to motion correction algorithms
Detecting Motion Artifacts and generating the MA mask
[7]:
# we use Optical Density data for motion artifact detection
fNIRSdata = rec["od"]
# define parameters for motion artifact detection. We follow the method from Homer2/3: "hmrR_MotionArtifactByChannel" and "hmrR_MotionArtifact".
t_motion = 0.5*units.s # time window for motion artifact detection
t_mask = 1.0*units.s # time window for masking motion artifacts (+- t_mask s before/after detected motion artifact)
stdev_thresh = 4.0 # threshold for standard deviation of the signal used to detect motion artifacts. Default is 50. We set it very low to find something in our good data for demonstration purposes.
amp_thresh = 5.0 # threshold for amplitude of the signal used to detect motion artifacts. Default is 5.
# to identify motion artifacts with these parameters we call the following function
ma_mask = quality.id_motion(fNIRSdata, t_motion, t_mask, stdev_thresh, amp_thresh)
# it hands us a boolean mask (xarray) of the input dimension, where False indicates a motion artifact at a given time point.
# show the masks data
ma_mask
/home/runner/micromamba/envs/cedalion/lib/python3.11/site-packages/xarray/core/variable.py:338: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
/home/runner/micromamba/envs/cedalion/lib/python3.11/site-packages/xarray/core/variable.py:338: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
data = np.asarray(data)
[7]:
<xarray.DataArray (channel: 28, wavelength: 2, time: 23239)> Size: 1MB
array([[[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]],
[[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]],
[[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]],
...,
[[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]],
[[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True]],
[[ True, True, True, ..., True, True, True],
[False, False, False, ..., True, True, True]]])
Coordinates:
* time (time) float64 186kB 0.0 0.128 0.256 ... 2.974e+03 2.974e+03
samples (time) int64 186kB 0 1 2 3 4 5 ... 23234 23235 23236 23237 23238
* channel (channel) object 224B 'S1D1' 'S1D2' 'S1D3' ... 'S8D8' 'S8D16'
source (channel) object 224B 'S1' 'S1' 'S1' 'S1' ... 'S8' 'S8' 'S8'
detector (channel) object 224B 'D1' 'D2' 'D3' 'D9' ... 'D7' 'D8' 'D16'
* wavelength (wavelength) float64 16B 760.0 850.0The output mask is quite detailed and still contains all original dimensions (e.g. single wavelengths) and allows us to combine it with a mask from another motion artifact detection method. This is the same approach as for the channel quality metrics above.
Let us now plot the result for an example channel. Note, that for both wavelengths a different number of artifacts was identified, which can sometimes happen:
[8]:
p.figure()
p.plot(ma_mask.sel(time=slice(0,250)).time, ma_mask.sel(channel="S3D3", wavelength="760", time=slice(0,250)), "b-")
p.plot(ma_mask.sel(time=slice(0,250)).time, ma_mask.sel(channel="S3D3", wavelength="850", time=slice(0,250)), "r-")
p.xlabel("time / s")
p.ylabel("Motion artifact mask")
p.show()
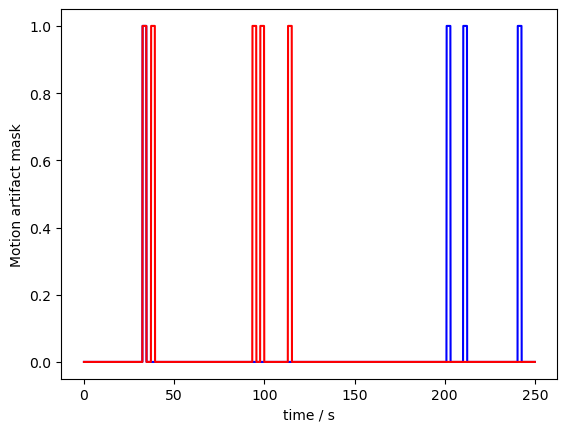
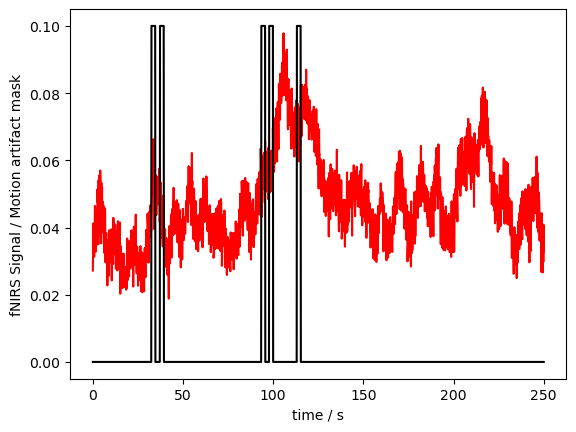
Our example dataset is very clean. So we artificially detected motion artifacts with a very low threshold. Plotting the mask and the data together (we have to rescale a bit to make both fit):
[9]:
p.figure()
p.plot(fNIRSdata.sel(time=slice(0,250)).time, fNIRSdata.sel(channel="S3D3", wavelength="760", time=slice(0,250)), "r-")
p.plot(ma_mask.sel(time=slice(0,250)).time, ma_mask.sel(channel="S3D3", wavelength="850", time=slice(0,250))/10, "k-")
p.xlabel("time / s")
p.ylabel("fNIRS Signal / Motion artifact mask")
p.show()
Refining the MA Mask
At the latest when we want to correct motion artifacts, we usually do not need the level of granularity that the mask provides. For instance, we usually want to treat a detected motion artifact in either of both wavelengths or chromophores of one channel as a single artifact that gets flagged for both. We might also want to flag motion artifacts globally, i.e. mask time points for all channels even if only some of them show an artifact. This can easily be done by using the “id_motion_refine” function. The function also returns useful information about motion artifacts in each channel in “ma_info”
[10]:
# refine the motion artifact mask. This function collapses the mask along dimensions that are chosen by the "operator" argument.
# Here we use "by_channel", which will yield a mask for each channel by collapsing the masks along either the wavelength or concentration dimension.
ma_mask_refined, ma_info = quality.id_motion_refine(ma_mask, 'by_channel')
# show the refined mask
ma_mask_refined
[10]:
<xarray.DataArray (channel: 28, time: 23239)> Size: 651kB
array([[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
...,
[ True, True, True, ..., True, True, True],
[ True, True, True, ..., True, True, True],
[False, False, False, ..., True, True, True]])
Coordinates:
* time (time) float64 186kB 0.0 0.128 0.256 ... 2.974e+03 2.974e+03
samples (time) int64 186kB 0 1 2 3 4 5 ... 23234 23235 23236 23237 23238
* channel (channel) object 224B 'S1D1' 'S1D2' 'S1D3' ... 'S8D8' 'S8D16'
source (channel) object 224B 'S1' 'S1' 'S1' 'S1' ... 'S7' 'S8' 'S8' 'S8'
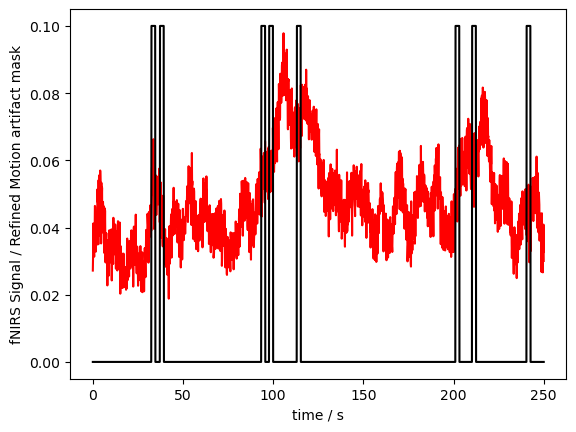
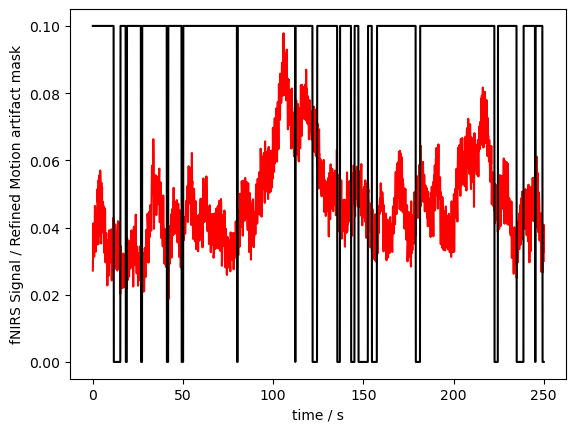
detector (channel) object 224B 'D1' 'D2' 'D3' 'D9' ... 'D7' 'D8' 'D16'Now the mask does not have the “wavelength” or “concentration” dimension anymore, and the masks of these dimensions are combined:
[11]:
# plot the figure
p.figure()
p.plot(fNIRSdata.sel(time=slice(0,250)).time, fNIRSdata.sel(channel="S3D3", wavelength="760", time=slice(0,250)), "r-")
p.plot(ma_mask_refined.sel(time=slice(0,250)).time, ma_mask_refined.sel(channel="S3D3", time=slice(0,250))/10, "k-")
p.xlabel("time / s")
p.ylabel("fNIRS Signal / Refined Motion artifact mask")
p.show()
# show the information about the motion artifacts: we get a pandas dataframe telling us
# 1) for which channels artifacts were detected,
# 2) what is the fraction of time points that were marked as artifacts and
# 3) how many artifacts where detected
ma_info
[11]:
| channel | ma_fraction | ma_count | |
|---|---|---|---|
| 0 | S1D1 | 0.097379 | 105 |
| 1 | S1D2 | 0.055854 | 64 |
| 2 | S1D3 | 0.040105 | 48 |
| 3 | S1D9 | 0.070141 | 89 |
| 4 | S2D1 | 0.022161 | 28 |
| 5 | S2D3 | 0.041439 | 47 |
| 6 | S2D4 | 0.026894 | 20 |
| 7 | S2D10 | 0.091484 | 100 |
| 8 | S3D2 | 0.013254 | 18 |
| 9 | S3D3 | 0.035888 | 46 |
| 10 | S3D11 | 0.095142 | 101 |
| 11 | S4D3 | 0.023925 | 25 |
| 12 | S4D4 | 0.042300 | 29 |
| 13 | S4D12 | 0.066440 | 58 |
| 14 | S5D5 | 0.081372 | 93 |
| 15 | S5D6 | 0.048023 | 57 |
| 16 | S5D7 | 0.044064 | 54 |
| 17 | S5D13 | 0.135075 | 126 |
| 18 | S6D5 | 0.031628 | 34 |
| 19 | S6D7 | 0.037652 | 46 |
| 20 | S6D8 | 0.037738 | 34 |
| 21 | S6D14 | 0.086751 | 87 |
| 22 | S7D6 | 0.062266 | 71 |
| 23 | S7D7 | 0.048152 | 52 |
| 24 | S7D15 | 0.122079 | 126 |
| 25 | S8D7 | 0.028831 | 34 |
| 26 | S8D8 | 0.030767 | 19 |
| 27 | S8D16 | 0.055037 | 52 |
Now we look at the “all” operator, which will collapse the mask across all dimensions except time, leading to a single motion artifact mask
[12]:
# "all", yields a mask that flags an artifact at any given time if flagged for any channel, wavelength, chromophore, etc.
ma_mask_refined, ma_info = quality.id_motion_refine(ma_mask, 'all')
# show the refined mask
ma_mask_refined
[12]:
<xarray.DataArray (time: 23239)> Size: 23kB
array([False, False, False, ..., True, True, True])
Coordinates:
* time (time) float64 186kB 0.0 0.128 0.256 ... 2.974e+03 2.974e+03
samples (time) int64 186kB 0 1 2 3 4 5 ... 23234 23235 23236 23237 23238[13]:
# plot the figure
p.figure()
p.plot(fNIRSdata.sel(time=slice(0,250)).time, fNIRSdata.sel(channel="S3D3", wavelength="760", time=slice(0,250)), "r-")
p.plot(ma_mask_refined.sel(time=slice(0,250)).time, ma_mask_refined.sel(time=slice(0,250))/10, "k-")
p.xlabel("time / s")
p.ylabel("fNIRS Signal / Refined Motion artifact mask")
p.show()
# show the information about the motion artifacts: we get a pandas dataframe telling us
# 1) that the mask is for all channels
# 2) fraction of time points that were marked as artifacts for this mask across all channels
# 3) how many artifacts where detected in total
ma_info
[13]:
| channel | ma_fraction | ma_count | |
|---|---|---|---|
| 0 | all channels combined | [0.31911872283661086, 0.3049614871552132] | [274, 229] |