Basic single trial fNIRS finger tapping classification
This notebook sketches the analysis of a finger tapping dataset with multiple subjects. A simple Linear Discriminant Analysis (LDA) classifier is trained to distinguish left and right fingertapping.
[1]:
import cedalion
import cedalion.nirs
from cedalion.datasets import get_multisubject_fingertapping_snirf_paths
import numpy as np
import xarray as xr
import matplotlib.pyplot as p
from sklearn.preprocessing import LabelEncoder
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
xr.set_options(display_max_rows=3, display_values_threshold=50)
np.set_printoptions(precision=4)
Loading raw CW-NIRS data from a SNIRF file
This notebook uses a finger-tapping dataset in BIDS layout provided by Rob Luke. It can can be downloaded via cedalion.datasets.
Cedalion’s read_snirf method returns a list of Recording objects. These are containers for timeseries and adjunct data objects.
[2]:
fnames = get_multisubject_fingertapping_snirf_paths()
subjects = [f"sub-{i:02d}" for i in [1, 2, 3, 4, 5]]
# store data of different subjects in a dictionary
data = {}
for subject, fname in zip(subjects, fnames):
records = cedalion.io.read_snirf(fname)
rec = records[0]
display(rec)
# Cedalion registers an accessor (attribute .cd ) on pandas DataFrames.
# Use this to rename trial_types inplace.
rec.stim.cd.rename_events(
{"1.0": "control", "2.0": "Tapping/Left", "3.0": "Tapping/Right"}
)
dpf = xr.DataArray(
[6, 6],
dims="wavelength",
coords={"wavelength": rec["amp"].wavelength},
)
rec["od"] = -np.log(rec["amp"] / rec["amp"].mean("time")),
rec["conc"] = cedalion.nirs.beer_lambert(rec["amp"], rec.geo3d, dpf)
data[subject] = rec
Downloading file 'multisubject-fingertapping.zip' from 'https://doc.ml.tu-berlin.de/cedalion/datasets/multisubject-fingertapping.zip' to '/home/runner/.cache/cedalion'.
Unzipping contents of '/home/runner/.cache/cedalion/multisubject-fingertapping.zip' to '/home/runner/.cache/cedalion/multisubject-fingertapping.zip.unzip'
<Recording | timeseries: ['amp'], masks: [], stim: ['1.0', '15.0', '2.0', '3.0'], aux_ts: [], aux_obj: []>
<Recording | timeseries: ['amp'], masks: [], stim: ['1.0', '15.0', '2.0', '3.0'], aux_ts: [], aux_obj: []>
<Recording | timeseries: ['amp'], masks: [], stim: ['1.0', '15.0', '2.0', '3.0'], aux_ts: [], aux_obj: []>
<Recording | timeseries: ['amp'], masks: [], stim: ['1.0', '15.0', '2.0', '3.0'], aux_ts: [], aux_obj: []>
<Recording | timeseries: ['amp'], masks: [], stim: ['1.0', '15.0', '2.0', '3.0'], aux_ts: [], aux_obj: []>
Illustrate the dataset of one subject
[3]:
display(data["sub-01"])
<Recording | timeseries: ['amp', 'od', 'conc'], masks: [], stim: ['control', '15.0', 'Tapping/Left', 'Tapping/Right'], aux_ts: [], aux_obj: []>
Frequency filtering and splitting into epochs
[4]:
for subject, rec in data.items():
# cedalion registers the accessor .cd on DataArrays
# to provide common functionality like frequency filters...
rec["conc_freqfilt"] = rec["conc"].cd.freq_filter(
fmin=0.02, fmax=0.5, butter_order=4
)
# ... or epoch splitting
rec["cfepochs"] = rec["conc_freqfilt"].cd.to_epochs(
rec.stim, # stimulus dataframe
["Tapping/Left", "Tapping/Right"], # select events
before=5, # seconds before stimulus
after=20, # seconds after stimulus
)
Plot frequency filtered data
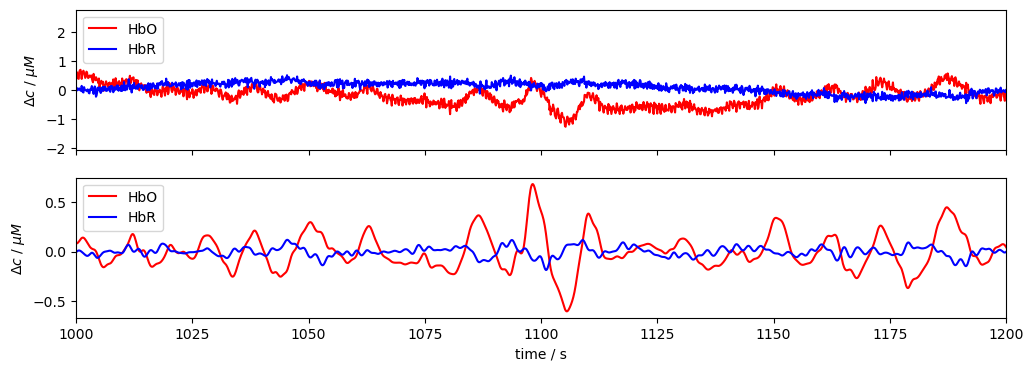
Illustrate for a single subject and channel the effect of the bandpass filter.
[5]:
rec = data["sub-01"]
channel = "S5D7"
f, ax = p.subplots(2, 1, figsize=(12, 4), sharex=True)
ax[0].plot(rec["conc"].time, rec["conc"].sel(channel=channel, chromo="HbO"), "r-", label="HbO")
ax[0].plot(rec["conc"].time, rec["conc"].sel(channel=channel, chromo="HbR"), "b-", label="HbR")
ax[1].plot(
rec["conc_freqfilt"].time,
rec["conc_freqfilt"].sel(channel=channel, chromo="HbO"),
"r-",
label="HbO",
)
ax[1].plot(
rec["conc_freqfilt"].time,
rec["conc_freqfilt"].sel(channel=channel, chromo="HbR"),
"b-",
label="HbR",
)
ax[0].set_xlim(1000, 1200)
ax[1].set_xlabel("time / s")
ax[0].set_ylabel("$\Delta c$ / $\mu M$")
ax[1].set_ylabel("$\Delta c$ / $\mu M$")
ax[0].legend(loc="upper left")
ax[1].legend(loc="upper left");
[6]:
display(data["sub-01"]["cfepochs"])
<xarray.DataArray 'concentration' (epoch: 60, chromo: 2, channel: 28,
reltime: 196)> Size: 5MB
<Quantity([[[[-1.9479e-02 -1.9950e-02 -2.0945e-02 ... -1.9397e-01 -2.1930e-01
-2.4552e-01]
[-1.0442e-02 -1.1375e-02 -1.2845e-02 ... -2.5376e-01 -2.5998e-01
-2.6454e-01]
[-1.2500e-02 -8.8897e-03 -5.8428e-03 ... -2.1148e-01 -2.2557e-01
-2.3975e-01]
...
[ 9.5644e-02 1.0075e-01 1.0499e-01 ... -3.2907e-01 -3.4702e-01
-3.6405e-01]
[ 2.0733e-02 2.2404e-02 2.3932e-02 ... -2.9462e-01 -2.9954e-01
-3.0371e-01]
[ 1.4324e-03 2.0415e-03 4.1027e-03 ... -3.0974e-01 -3.2710e-01
-3.4287e-01]]
[[ 1.4641e-02 6.4301e-03 -1.6853e-03 ... -6.5363e-02 -6.2839e-02
-5.7220e-02]
[ 2.0950e-03 5.5313e-03 8.9983e-03 ... -4.8858e-02 -5.4990e-02
-6.0070e-02]
[ 4.5053e-02 4.2005e-02 3.9300e-02 ... -4.3936e-02 -4.3039e-02
-4.0861e-02]
...
[ 2.8454e-01 2.9324e-01 3.0016e-01 ... 1.0617e-01 1.1339e-01
1.2119e-01]
[ 3.4306e-01 3.9592e-01 4.4809e-01 ... -1.8578e-01 -1.7872e-01
-1.7142e-01]
[ 6.1801e-01 6.1059e-01 6.0291e-01 ... 1.8868e-01 1.9003e-01
1.9076e-01]]
[[ 6.5167e-02 5.7419e-02 4.9134e-02 ... 3.1521e-02 3.2053e-02
3.1760e-02]
[ 3.6845e-02 3.4700e-02 3.1450e-02 ... -3.0447e-02 -2.7677e-02
-2.3868e-02]
[ 4.8888e-02 3.9980e-02 3.2743e-02 ... 3.0387e-02 3.4824e-02
3.9630e-02]
...
[ 5.8174e-02 5.3107e-02 4.7739e-02 ... -8.1364e-03 -6.2961e-03
-5.3040e-03]
[ 1.5154e-01 1.6202e-01 1.7299e-01 ... 2.6186e-02 3.2992e-02
3.8859e-02]
[ 1.5938e-01 1.5348e-01 1.4687e-01 ... 3.8222e-02 4.2833e-02
4.6774e-02]]]], 'micromolar')>
Coordinates: (3/6)
* chromo (chromo) <U3 24B 'HbO' 'HbR'
* channel (channel) object 224B 'S1D1' 'S1D2' 'S1D3' ... 'S8D8' 'S8D16'
... ...
trial_type (epoch) object 480B 'Tapping/Left' ... 'Tapping/Right'
Dimensions without coordinates: epoch[7]:
all_epochs = xr.concat([rec["cfepochs"] for rec in data.values()], dim="epoch")
all_epochs
[7]:
<xarray.DataArray 'concentration' (epoch: 300, chromo: 2, channel: 28,
reltime: 196)> Size: 26MB
<Quantity([[[[-1.9479e-02 -1.9950e-02 -2.0945e-02 ... -1.9397e-01 -2.1930e-01
-2.4552e-01]
[-1.0442e-02 -1.1375e-02 -1.2845e-02 ... -2.5376e-01 -2.5998e-01
-2.6454e-01]
[-1.2500e-02 -8.8897e-03 -5.8428e-03 ... -2.1148e-01 -2.2557e-01
-2.3975e-01]
...
[ 9.5644e-02 1.0075e-01 1.0499e-01 ... -3.2907e-01 -3.4702e-01
-3.6405e-01]
[ 2.0733e-02 2.2404e-02 2.3932e-02 ... -2.9462e-01 -2.9954e-01
-3.0371e-01]
[ 1.4324e-03 2.0415e-03 4.1027e-03 ... -3.0974e-01 -3.2710e-01
-3.4287e-01]]
[[ 1.4641e-02 6.4301e-03 -1.6853e-03 ... -6.5363e-02 -6.2839e-02
-5.7220e-02]
[ 2.0950e-03 5.5313e-03 8.9983e-03 ... -4.8858e-02 -5.4990e-02
-6.0070e-02]
[ 4.5053e-02 4.2005e-02 3.9300e-02 ... -4.3936e-02 -4.3039e-02
-4.0861e-02]
...
[-1.8642e-01 -1.8383e-01 -1.8031e-01 ... 9.3582e-03 1.1076e-02
1.3423e-02]
[-2.8335e-01 -2.7513e-01 -2.6600e-01 ... -1.3274e-02 -4.6116e-03
2.3699e-03]
[-3.7102e-01 -3.6417e-01 -3.5820e-01 ... 1.8997e-01 1.9610e-01
2.0283e-01]]
[[ 6.9413e-02 6.7628e-02 6.5086e-02 ... 2.3354e-02 8.2493e-03
-6.5091e-03]
[ 9.3748e-02 7.1599e-02 4.8922e-02 ... 6.0441e-03 3.1716e-02
4.8579e-02]
[ 1.3260e-01 1.2931e-01 1.2489e-01 ... 1.8629e-01 1.6657e-01
1.4594e-01]
...
[ 3.5046e-02 3.1738e-02 2.8594e-02 ... 9.8597e-02 1.0093e-01
1.0222e-01]
[ 8.7634e-02 8.3598e-02 8.0126e-02 ... 1.1028e-01 1.0263e-01
9.5370e-02]
[-3.3306e-02 -3.6382e-02 -3.7580e-02 ... 4.1337e-03 3.1121e-03
5.3934e-04]]]], 'micromolar')>
Coordinates: (3/6)
* chromo (chromo) <U3 24B 'HbO' 'HbR'
* channel (channel) object 224B 'S1D1' 'S1D2' 'S1D3' ... 'S8D8' 'S8D16'
... ...
trial_type (epoch) object 2kB 'Tapping/Left' ... 'Tapping/Right'
Dimensions without coordinates: epochBlock Averages
[8]:
# calculate baseline
baseline = all_epochs.sel(reltime=(all_epochs.reltime < 0)).mean("reltime")
# subtract baseline
all_epochs_blcorrected = all_epochs - baseline
# group trials by trial_type. For each group individually average the epoch dimension
blockaverage = all_epochs_blcorrected.groupby("trial_type").mean("epoch")
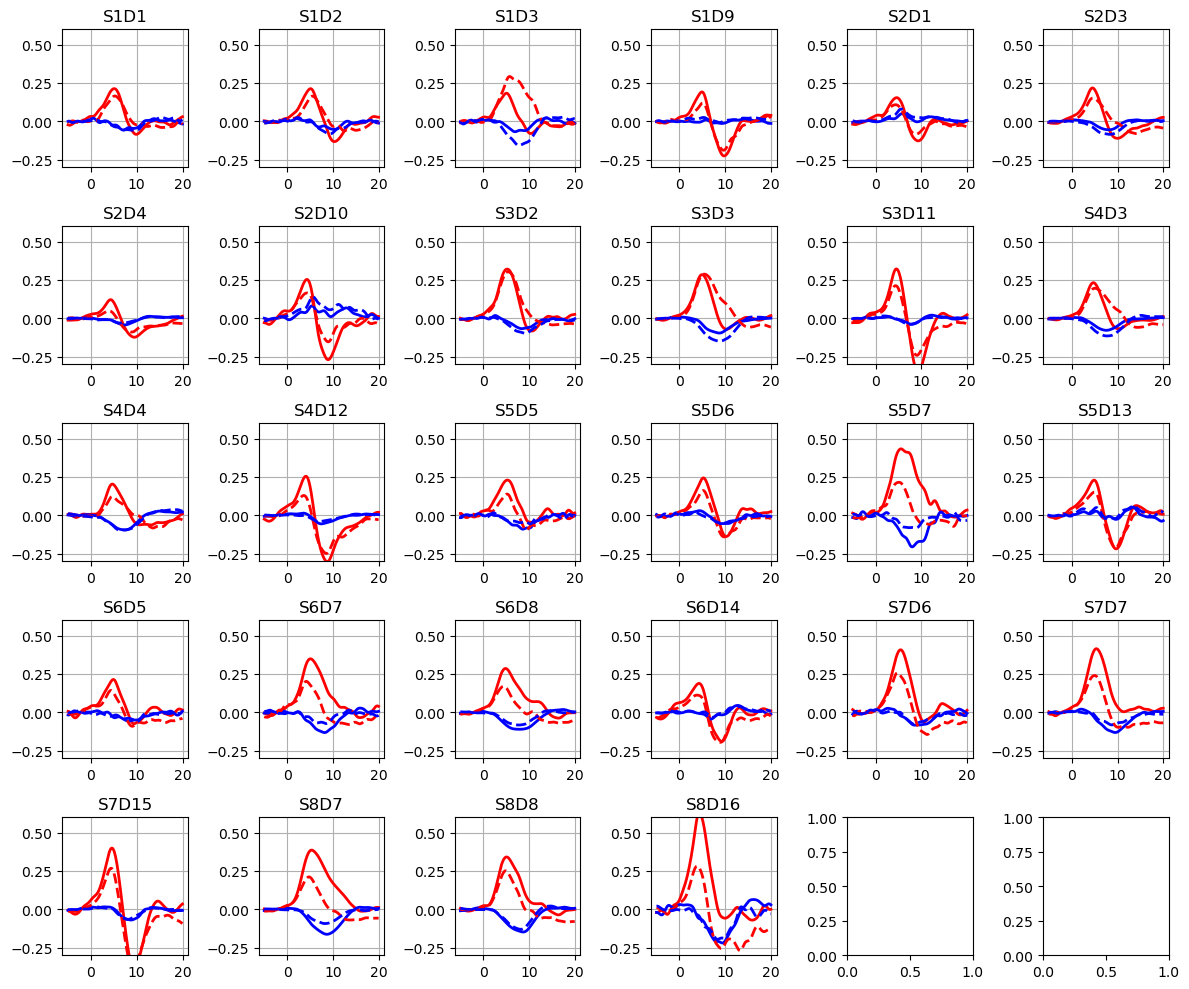
Plotting averaged epochs
[9]:
f, ax = p.subplots(5, 6, figsize=(12, 10))
ax = ax.flatten()
for i_ch, ch in enumerate(blockaverage.channel):
for ls, trial_type in zip(["-", "--"], blockaverage.trial_type):
ax[i_ch].plot(
blockaverage.reltime,
blockaverage.sel(chromo="HbO", trial_type=trial_type, channel=ch),
"r",
lw=2,
ls=ls,
)
ax[i_ch].plot(
blockaverage.reltime,
blockaverage.sel(chromo="HbR", trial_type=trial_type, channel=ch),
"b",
lw=2,
ls=ls,
)
ax[i_ch].grid(1)
ax[i_ch].set_title(ch.values)
ax[i_ch].set_ylim(-0.3, 0.6)
p.tight_layout()
Training a LDA classifier with Scikit-Learn
[10]:
# start with the frequency-filtered, epoched and baseline-corrected concentration data
# discard the samples before the stimulus onset
epochs = all_epochs_blcorrected.sel(reltime=all_epochs_blcorrected.reltime >=0)
# strip units. sklearn would strip them anyway and issue a warning about it.
epochs = epochs.pint.dequantify()
# need to manually tell xarray to create an index for trial_type
epochs = epochs.set_xindex("trial_type")
[11]:
display(epochs)
<xarray.DataArray 'concentration' (epoch: 300, chromo: 2, channel: 28,
reltime: 157)> Size: 21MB
array([[[[-3.6270e-02, -1.8064e-02, 2.0471e-03, ..., -6.8976e-02,
-9.4304e-02, -1.2053e-01],
[ 4.6475e-03, 1.1837e-02, 1.9097e-02, ..., -1.5186e-01,
-1.5808e-01, -1.6264e-01],
[ 2.4496e-03, 1.2294e-02, 2.3849e-02, ..., -1.3250e-01,
-1.4658e-01, -1.6077e-01],
...,
[-4.3450e-02, -3.0909e-02, -1.6201e-02, ..., -3.4983e-01,
-3.6779e-01, -3.8482e-01],
[ 2.2952e-02, 3.4524e-02, 4.6622e-02, ..., -2.6465e-01,
-2.6958e-01, -2.7375e-01],
[-4.2557e-03, 1.3953e-02, 3.3226e-02, ..., -2.5881e-01,
-2.7617e-01, -2.9194e-01]],
[[ 1.5914e-02, 1.3944e-02, 1.0925e-02, ..., -7.7108e-02,
-7.4584e-02, -6.8965e-02],
[-1.2916e-02, -9.4326e-03, -4.7285e-03, ..., -5.5005e-02,
-6.1136e-02, -6.6217e-02],
[-6.1391e-03, -2.1726e-03, -2.9566e-05, ..., -8.1372e-02,
-8.0476e-02, -7.8297e-02],
...
[ 1.5353e-01, 1.5630e-01, 1.5688e-01, ..., 6.7293e-02,
6.9011e-02, 7.1357e-02],
[ 2.4956e-01, 2.4903e-01, 2.4447e-01, ..., 7.8465e-02,
8.7128e-02, 9.4109e-02],
[ 4.8391e-01, 4.8248e-01, 4.7416e-01, ..., 3.1090e-01,
3.1703e-01, 3.2376e-01]],
[[-1.5826e-02, -1.2935e-02, -5.2803e-03, ..., 2.3303e-03,
-1.2774e-02, -2.7533e-02],
[-1.3622e-02, -1.8221e-02, -2.3395e-02, ..., 1.1320e-02,
3.6992e-02, 5.3854e-02],
[-3.5104e-02, -3.3747e-02, -3.9994e-02, ..., 7.3406e-02,
5.3683e-02, 3.3055e-02],
...,
[ 1.1524e-02, 1.4339e-02, 1.6261e-02, ..., 9.8163e-02,
1.0049e-01, 1.0178e-01],
[-5.6057e-02, -5.8393e-02, -5.9799e-02, ..., 6.0866e-02,
5.3214e-02, 4.5954e-02],
[ 3.0087e-02, 3.2592e-02, 3.6472e-02, ..., -5.2316e-03,
-6.2531e-03, -8.8259e-03]]]])
Coordinates: (3/6)
* chromo (chromo) <U3 24B 'HbO' 'HbR'
* channel (channel) object 224B 'S1D1' 'S1D2' 'S1D3' ... 'S8D8' 'S8D16'
... ...
* trial_type (epoch) object 2kB 'Tapping/Left' ... 'Tapping/Right'
Dimensions without coordinates: epoch
Attributes:
units: micromolar[12]:
X = epochs.stack(features=["chromo", "channel", "reltime"])
display(X)
<xarray.DataArray 'concentration' (epoch: 300, features: 8792)> Size: 21MB
array([[-0.0363, -0.0181, 0.002 , ..., -0.0126, -0.0093, -0.0072],
[ 0.109 , 0.0983, 0.0847, ..., -0.0343, -0.036 , -0.0378],
[-0.0203, -0.0267, -0.0325, ..., -0.0187, -0.0155, -0.0115],
...,
[ 0.029 , 0.0078, -0.0164, ..., -0.0117, -0.0013, 0.0085],
[-0.2281, -0.2284, -0.2273, ..., 0.0104, 0.0108, 0.0135],
[ 0.2505, 0.2608, 0.2643, ..., -0.0052, -0.0063, -0.0088]])
Coordinates: (3/7)
source (features) object 70kB 'S1' 'S1' 'S1' 'S1' ... 'S8' 'S8' 'S8'
detector (features) object 70kB 'D1' 'D1' 'D1' 'D1' ... 'D16' 'D16' 'D16'
... ...
* reltime (features) float64 70kB 0.0 0.128 0.256 ... 19.71 19.84 19.97
Dimensions without coordinates: epoch
Attributes:
units: micromolar[13]:
y = xr.apply_ufunc(LabelEncoder().fit_transform, X.trial_type)
display(y)
<xarray.DataArray 'trial_type' (epoch: 300)> Size: 2kB
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1])
Coordinates:
* trial_type (epoch) object 2kB 'Tapping/Left' ... 'Tapping/Right'
Dimensions without coordinates: epoch[14]:
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, stratify=y)
classifier = LinearDiscriminantAnalysis(n_components=1).fit(X_train, y_train)
y_pred = classifier.predict(X_test)
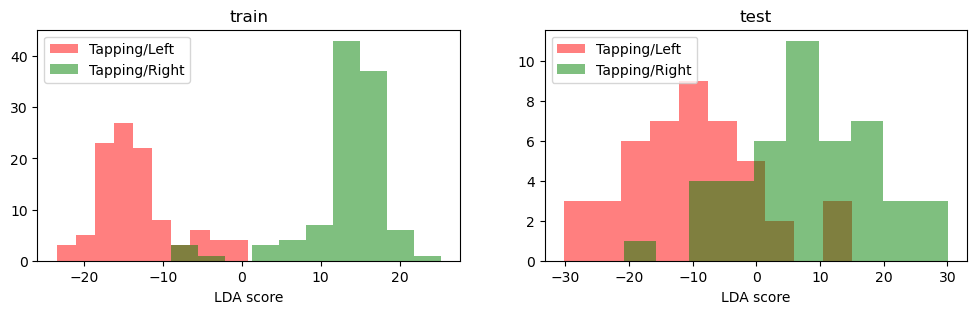
print(f"Accuracy: {accuracy_score(y_test, y_pred)}")
Accuracy: 0.8888888888888888
[15]:
f, ax = p.subplots(1, 2, figsize=(12, 3))
for trial_type, c in zip(["Tapping/Left", "Tapping/Right"], ["r", "g"]):
kw = dict(alpha=0.5, fc=c, label=trial_type)
ax[0].hist(classifier.decision_function(X_train.sel(trial_type=trial_type)), **kw)
ax[1].hist(classifier.decision_function(X_test.sel(trial_type=trial_type)), **kw)
ax[0].set_xlabel("LDA score")
ax[1].set_xlabel("LDA score")
ax[0].set_title("train")
ax[1].set_title("test")
ax[0].legend(ncol=1, loc="upper left")
ax[1].legend(ncol=1, loc="upper left");